nf-core/tumourevo
Analysis pipeline to model tumour clonal evolution from WGS data (driver annotation, quality control of copy number calls, subclonal and mutational signature deconvolution)
Introduction
nf-core/tumourevo is a bioinformatics pipeline to model tumour evolution from whole-genome sequencing (WGS) data. The pipeline performs state-of-the-art downstream analysis of variant and copy-number calls from tumour-normal matched sequencing assays, reconstructing the evolutionary processes leading to the observed tumour genome. This analysis can be done at the level of single samples, multiple samples from the same patient (multi-region/longitudinal assays), and of multiple patients from distinct cohorts.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
Pipeline Summary
The tumourevo pipeline supports variant annotation, driver annotation, quality control processes, subclonal deconvolution and signature deconvolution analysis through various tools. It can be used to analyse both single sample experiments and longitudinal/multi-region assays, in which multiple samples of the same patient are avaiable. As input, you must provide at least information on the samples, the VCF file from one of the supported callers and the output of one of the supported copy number caller. By default, if multiple samples from the same patient are provided, they will be analysed in a multivariate framework (which affects in particular the subclonal deconvolution deconvolution steps) to retrieve information useful in the reconstruction of the evolutionary process. Depending on the variant calling strategy (single sample or multi sample) and the provided input files, different strategies will be applied.
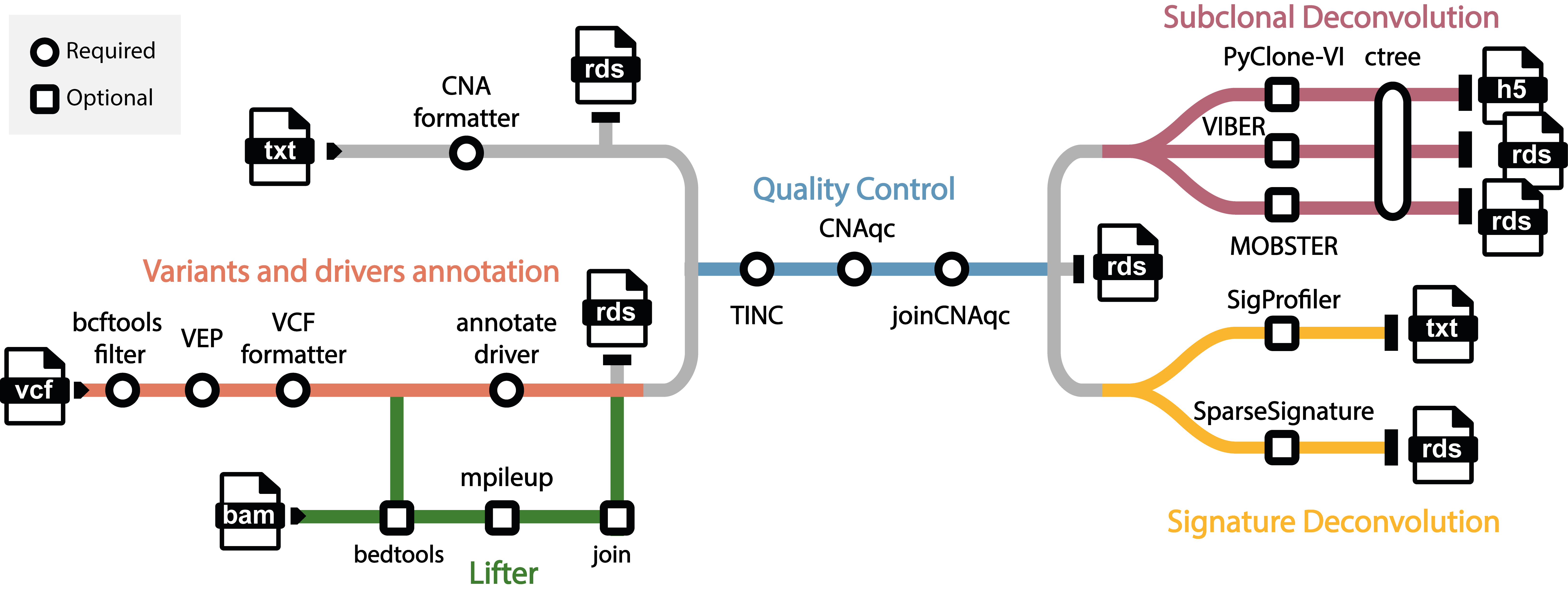
- Variant Annotation (
VEP) - Quality Control (
CNAqc,TINC) - Driver Annotation
- Subclonal Deconvolution (
PyClone,MOBSTER,VIBER) - Clone Tree Inference (
ctree) - Signature Deconvolution (
SparseSignatures,SigProfiler) - Genome Interpreter
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
dataset,patient,tumour_sample,normal_sample,vcf,tbi,cna_segments,cna_extra,cna_caller,cancer_type
dataset1,patient1,sample1,N1,patient1_sample1.vcf.gz,patient1_sample1.vcf.gz.tbi,/CNA/patient1/sample1/segments.txt,CNA/patient1/sample1/purity_ploidy.txt,caller,PANCANCER
dataset1,patient1,sample2,N1,patient1_sample2.vcf.gz,patient1_sample2.vcf.gz.tbi,/CNA/patient1/sample2/segments.txt,CNA/patient1/sample2/purity_ploidy.txt,caller,PANCANCERNow, you can run the pipeline using:
nextflow run nf-core/tumourevo \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
--outdir <OUTDIR> \
--genome GRCh37 \
--fasta <PATH>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any
configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/tumourevo was originally written by Nicola Calonaci, Elena Buscaroli, Katsiaryna Davydzenka, Giorgia Gandolfi, Virginia Gazziero, Brandon Hastings, Davide Rambaldi, Rodolfo Tolloi, Lucrezia Valeriani and Giulio Caravagna.
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #tumourevo channel (you can join with this invite).
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.